WHAT'S NEW?
This is SIREs Version 3.0
31/01/2024
NEW INPUT MODES: We have included 2 new modes that automatically query NCBI and Ensembl databases by Gene Name or Transcript ID to obtain the sequences and predict IREs on them with only one click. This version also includes a Batch mode enabling larger submissions up to thousands of sequences and millions of residues, to predict IREs. The batch server provides a list of predicted IREs in tabular format to browse the results. From the browsable table you can select up to 10 sequences for direct submission to the interactive server where you will get the complete output.
IRE LOCATION IN TRANSCRIPT: SIREs predictions now contain the location of the predicted element: 5'UTR, 5'UTR-CDS, CDS, CDS-3'UTR, 3'UTR or Intron.
IREs IN CONTEXT: In addition to the quality score that our program provides, we now include a graphical element to compare the scores and free-energy of predicted IREs with curated set of canonical IREs from various species. This visual reference aims guide researchers in identifying candidates worthy of further study.
SCORING SYSTEM MODIFICATION: We have also improved the scoring system, to account for more fine-grained sequence- and structural-related details.
NOVEL MOTIFS: We have added 4 new motifs (19 to 22) to increase the predictive power of SIREs. Check the documentation to learn more about them.
EXTENDED DOCUMENTATION: We have updated the FAQS, Documentation and References pages to include more up-to-date information. We have also included a Troubleshooting page to help users understand potential error messages during their use of the SIREs server. Additionally, we also included this page to keep track of the novelties included on every SIREs version.
PYTHON MIGRATION: We have migrated the code from Perl to Python, aiming at guaranteing an easier maintenance in the future and integration with external software and state-of-the-art libraries and packages.
ViennaRNA PACKAGE UPDATE: We have also updated ViennaRNA package to the last stable version (2.6.4). This update may change a little your previous results.
RENEWED INTERFACE: Finally, we have a new interface! We have worked to provide a friendly user-experience more concordant with the current times.
Previous versions
SIREs Version 2.1 - 10/03/2015
ViennaRNA PACKAGE UPDATE: We updated the software to version (2.1.9). This update may change a little your previous results.
SIREs Version 2.0 - 31/12/2012

REFERENCE SECTION UPDATED: The Reference section now includes a link to the list of papers that cite the SIRES NAR paper.
FAQ SECTION UPDATED: We have updated the FAQ Section to include this information.
SIREs Version 1.0 - 2010
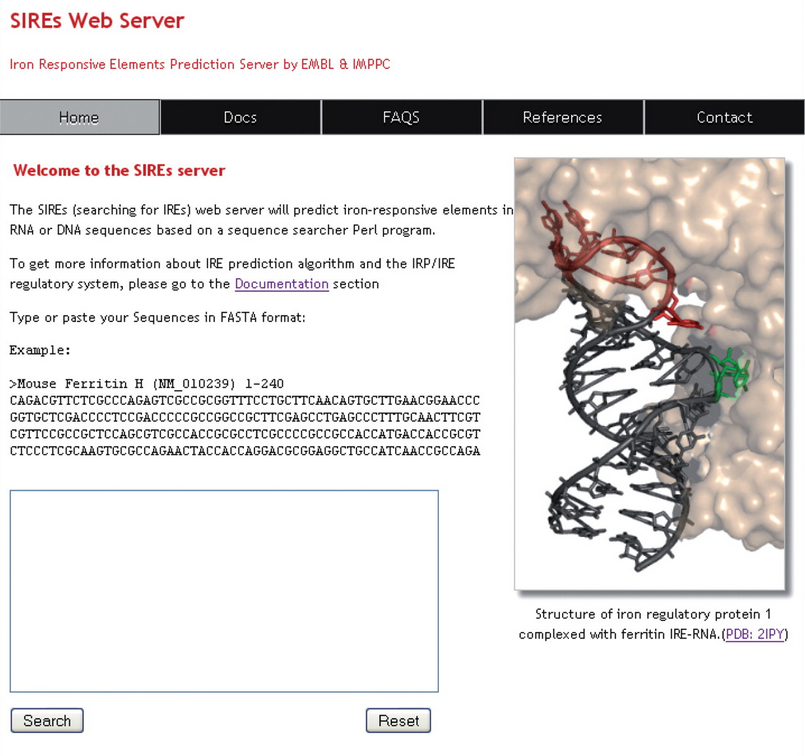
PERL-BASED PROGRAM: SIREs is based on a Perl script that screens for a 19 or 20 nucleotide sequence motif corresponding to the core sequence of an IRE (positions N7–N25) that includes the hexa-apical hairpin loop (N14–N19), the upper stem, the cytosine bulge (C8) and the lower base pair (N7–N25)
INTERACTIVE MODE: The server can accept single or multiple DNA or RNA sequence entries in FASTA format. For performance reasons, there is a limit of 500 000 residues to be submitted.
18 MOTIFS: These 18 motifs are based on two canonical IRE motifs 5'-CXXXXXCAGUGX-3' (motif 1) and 5'-CXXXXXCAGAGX-3' (motif 2) and 16 SELEX (systematic evolution of ligands by exponential enrichment) motifs proven to bind IRP1 and/or IRP2 in vitro with a relative binding efficiency >20%
CONFIDENCE SCORE: IREs are classified into three categories of confidence according to the sum score value: high (scores from 8 to 6); medium (scores from 5.9 to 4) and low (scores from 3.9 to 0). Read more about the scoring system in the documentation section.
RESULTS: Results are reported in tabular format and by a schematic visual representation that highlights important features of the IRE motif in color
ViennaRNA PACKAGE: SIREs uses RNAFold from the Vienna web suite (version 1.X) to fold the putative IRE and obtain the free energy of the structure.

